Clustering Book editions
using Python to increase user experience and data quality by clustering book editions

What do the books “The invention of Nature” and “De uitvinder van de natuur” have in common? Well, they are both different versions of the same work “The invention of nature” by Andrea Wulf, whether it is in a different format or a different language. In this blog post I will briefly introduce the advantages of keeping work-level records in library catalogs. Furthermore, I will introduce a fast Python implementation (DOI: 10.5281/zenodo.10011416) which we used in the BELTRANS project to identify the works in a corpus of book translations #FRBRization.
Context
Library catalogs usually contain records about different versions of a book. In technical terms, these records are so-called manifestations, but for simplicity I will refer to them simply as book versions or book editions. Such a conceptual division between different levels of description is part of the Functional Requirements for Bibliographic Records (FRBR) or how it is referred to nowadays, the IFLA Library Reference Model (IFLA-LRM).
Better user experience and better data quality due to identified works
Imagine that you are searching for a specific book in a library catalog. You remember that it had the word “invention” in its title. Instead of seeing two search results “The invention of nature” by Andrea Wulf and “Invention: A Life of Learning through Failure” by James Dyson, you are overwhelmed with hundreds of versions of both books and others such as ebooks, paperbacks, reprints, etc. Actually you are interested in finding out which works the library has that contain the title. Which specific version maybe is of less priority to you. In such a scenario it might be better to only show works in the search results. After clicking on the work you looked for, you still can be presented with all the different versions of the book.
There is also an advantage for the library personnel that works with the book data. Creating catalog records often is a manual process, or at least it was for a long time. Some of the records might contain more information than others, for example the genre according to a specific categorization. Identifying all the different versions of a book in your own catalog or in a collection of (other) catalogs can help to fill gaps in the data. If one book record is classified as a Comic and mentions a specific person as its illustrator, another record of a different version of that book which maybe has no such information, can be completed/enriched with this information.
Alright, you convinced me, how can I do it?
There are different ways to do this. I have read a bit of literature and it seems that there is one very common way to do this. Let me introduce you to the work-set clustering algorithm, initially devised by OCLC. The idea is simple: create several possible descriptive keys of a book version based on a combination of work-level information such as title and author, then check if two book versions have at least one key in common. Sounds complicated? Check out the image below which hopefully makes it clear.
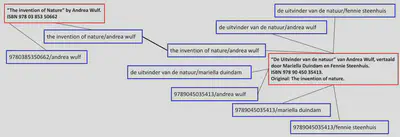
In this example you can see two book records (red), each describing a book version. For each book version descriptive keys (blue) are generated to assist in finding matches. As you can see, both records have at least one key in common. We found a match! In this case only because we also considered a possible original title as part of the key. Similarly we would find a match between the shown Dutch version of the printed book and a hypothetical ebook version: the ISBN would be different, but there would be a match based on the title-author or title-translator combination.
Nice idea, how do I write a program to do this?
That’s the one million dollar question. Different scientific papers talk about the algorithm, a few share code. But a code implementation always depends on the context it was written for, for example by using XSLT rules for XML records. I tried to write a program in Python that is as generic as possible and came up with three solutions. Why three? Well, it turned out the first two were not fast enough for the amount of translations we have in the BELTRANS project.
First attempt: reusing the Python package sklearn
Most of the papers I have read are already a few years old,
they mainly use software stacks that are less common nowadays.
I needed a solution that I can quickly run without a large setup.
Currently a lot of code libraries for common languages such as Python exist.
So why not reuse an existing one?!
The class AgglomerativeClustering from the Python package sklearn implements a hierarchical clustering algorithm.
Actually I am not interested in different levels of clusters, only the highest level with the least number of clusters. So I did what a lot of people do nowadays, I had a chat with ChatGPT to brainstorm possible solutions with AgglomerativeClustering. And it came up with a smart idea!
The input for the clustering is a so-called distance matrix: basically a table listing all elements as rows and all elements as columns, where the value in a cell indicates how similar an element in row X is with the element in column Y. We don’t need a sophisticated similarity measure, a simple “yes” for an overlap would suffice.
ChatGPTs idea was to use negative distance values. Like this, the whole clustering just takes two lines of code where I instruct the AgglomerativeClustering library to return clusters with distance threshold zero.
from sklearn.cluster import AgglomerativeClustering
model = AgglomerativeClustering(n_clusters=None, \
affinity='precomputed', \
linkage='single', \
distance_threshold=0)
# get a list where each index
# corresponds to an element in elementIDs
# e.g. [0,2,0,1] the first element is in cluster 0,
# the second in cluster 2,
# the third in cluster 0 and the 4th in cluster 1
clusterLabels = model.fit_predict(distanceMatrix)
This went well for a relevant subset of roughly 20 thousand records in our BELTRANS project. After a few minutes we got correct and directly reusable results! However, when we wanted to achieve the same for more than 150 thousand records, this implementation of the clustering was not performant enough. Even on a powerful server with more than 32 GB of RAM, the computation of the distance matrix takes too much time and space.
AgglomerativeClustering module does not support this.
Second attempt: implementing the algorithm myself
A little disappointed from the last solution, I thought I will try implementing it myself, all I need: a few data structures and loops. The idea of the algorithm is to start with every element in its own cluster. Then comparing it to other clusters until we find a match. When a match is found the loop is stopped, the clusters are merged and we loop again over the (now updated) list of clusters. We do all this until no more merges are possible.
This algorithm does not need a large distance matrix to start with. Testing if there is an overlap between two clusters can be reduced to a single intersection operation between two Python sets that represent the descriptive keys of the respective clusters.
Unfortunately all that takes too long. With a few example data I could verify that it worked as expected, but as soon as there are a few thousand elements the algorithm keeps running …
ThreadPoolExecutor or ProcessPoolExecutor, but it did not help much.
Third attempt: inverted index for clustering in no time
Even more frustrated I went back to the drawing board, or pen and paper in my case. What I need is a solution that ideally only runs once over all elements that need to be clustered. But how do I find out possible overlaps without comparing everything?
The disadvantages of the previous solution were that I had to perform set intersections to detect overlaps while iterating over all clusters, stop the iteration to update the clusters and start iterating again.
Actually the data already includes the matches between elements implicitly, and I used this to compute clusters in no time. Every element has one or more descriptive keys. But instead of only storing the mapping element->descriptive keys in a Python dictionary, I also store descriptive key->elements in another dictionary. This is my inverted index which I can use to look up overlaps.
Eventually I start with zero clusters and I simply iterate over the inverted index. If one of the elements from a key already is part of a cluster, I add the other elements to it. If no cluster was found, I create a new one and add the elements. Basically all elements of a descriptive key I check, either go to an existing cluster or end up in a new cluster. If different elements for a descriptive key are already in different clusters, a new merged cluster is created and the old ones deleted.
This solution works as well, and wow, it is fast too. Interestingly it takes longer to upload the computed cluster information to our database than performing the clustering itself. Inverted indexes can do a lot of heavy lifting!
You can find the source code on GitHub (DOI: 10.5281/zenodo.10011416).
Why is it so fast?
It all has to do with scalability. I only iterate once over all elements to build the inverted index dictionary. Then I iterate once over all elements in the inverted index to compute the clusters. For 100 input elements this roughly means iterating 200 times, for 100,000 input elements roughly 200,000 times. An algorithm that scales linear to its input.
Compare this to compute a distance matrix of 100*100 elements (compute and store 10 thousand results) and 100,000*100,000 elements (compute and store 10 billion results). An algorithm that scales quadratic to its input.
Final remarks
Having work-level information brings advantages for both librarians and users. The presented solution is just one possible implementation of one possible algorithm. Which data fields you use to create descriptive keys and how you extract them from your bibliographic data explicitly was not covered in this post. I also did not cover how you could use the information of the clusters to actually achieve the advantages. The main purpose of this blog post was to introduce and share the generic Python implementation, because I had the feeling there is not much out there that can be reused easily.
For the BELTRANS project we extracted descriptive keys with a SPARQL query from an Resource Description Framework (RDF) representation of bibliographic information. Similarly, we used SPARQL INSERT queries to explicitly create RDF representations for each work cluster and link its manifestations to it using the fabio ontology. Here you can have a look at our SPARQL queries.
Curious about more content like this or some behind-the-scenes? Consider subscribing to my bi-weekly Newsletter FAIR Data Digest to receive more interesting content about Linked Data every other Tuesday!
